Our research
The laboratory is devoted to the study of cell and cytoskeleton dynamics during early mammalian embryo development. We have two main themes of research: the mechanisms of gastrulation Epithelial-Mesenchymal Transition at the Primitive Streak after establishment of the anterior-posterior axis, and the subsequent morphogenesis of mesoderm-derived structures, in particular the extra-embryonic support membranes.
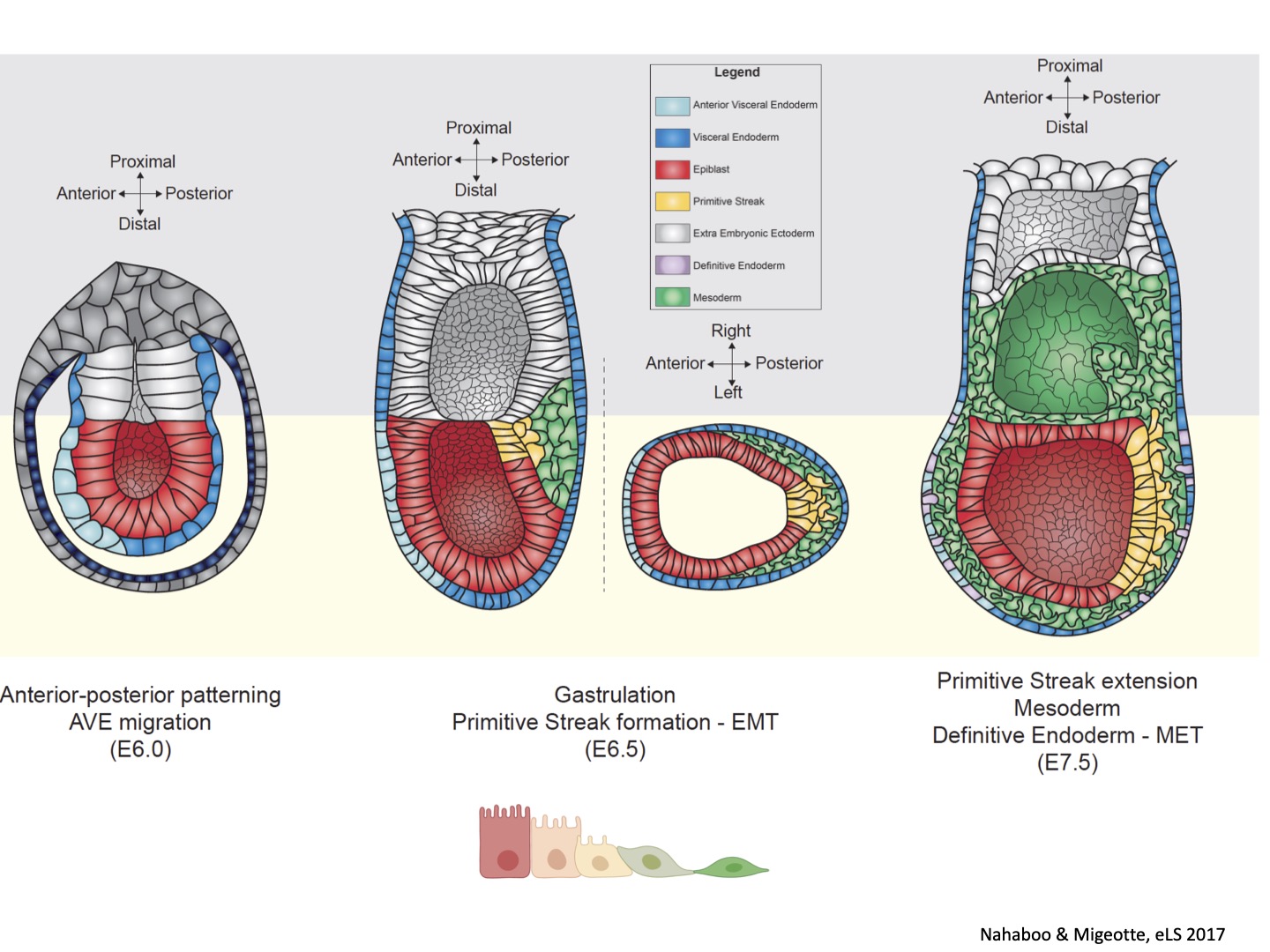
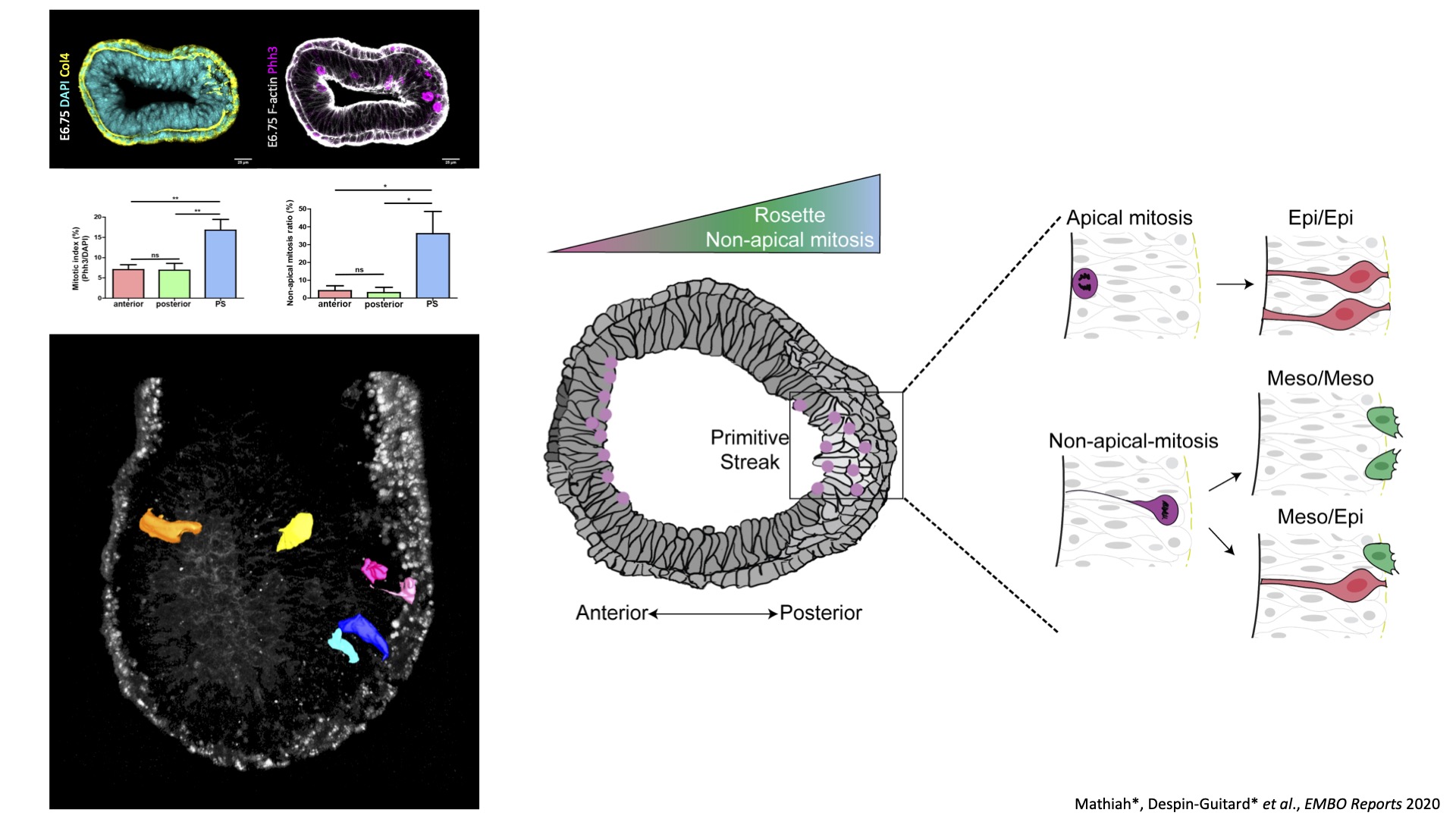
In collaboration with University of Cambridge, we participated to a study regarding the relationship between Anterior Visceral Endoderm migration and basal membrane perforation at the posterior side of the embryo at the initiation of gastrulation. We developed tools to observe epithelial-mesenchymal migration (EMT) at the Primitive Streak prior to mesoderm formation, and identified an asymmetry in mitosis frequency and position, suggesting a role for cell cycle in cell delamination. Current projects aim to better understand the mechanisms implicated in epithelial destabilization during gastrulation EMT, notably the implication of interkinetic nuclear migration, mitosis, and basal membrane degradation. We use mouse genetics and pharmacological screens, coupled with fixed and live imaging of embryos and stem cells-based embryo models.
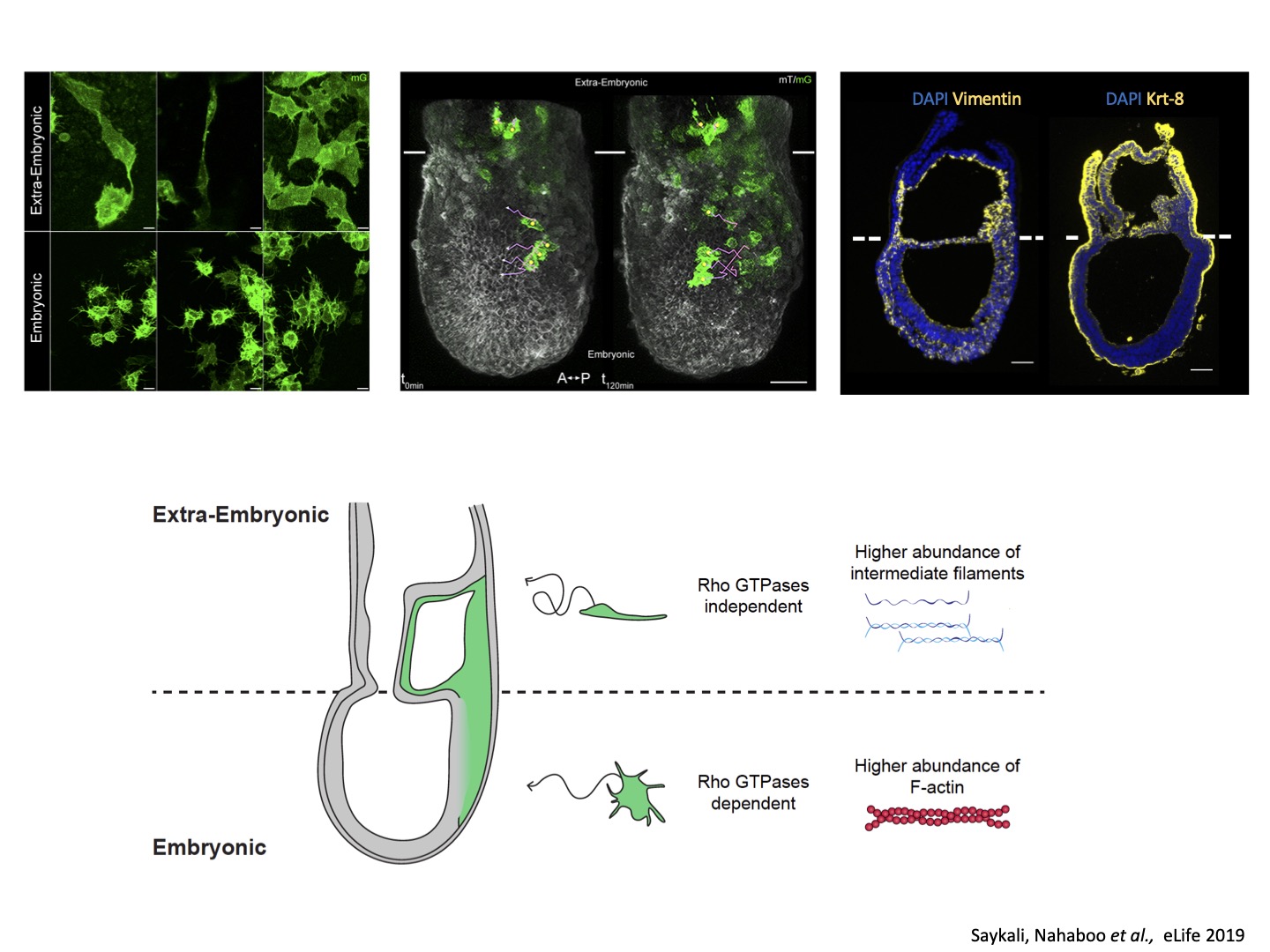
The lab has established original techniques for culture and live imaging of post-implantation mouse embryos, as well as mesoderm explants. Mechanistically, we focus on the role of cytoskeleton and mechanical forces. We identified differences in the patterns and mechanisms of mesoderm migration depending on whether mesoderm cells formed embryonic tissues or migrated to participate to extra-embryonic envelopes such as the amnion, umbilical cord, or placenta. We then identified a role for keratin intermediate filaments in the morphogenesis of those membranes.
Ongoing projects aim to:
- further define the interplay between cells’ cytoskeleton/adhesion repertoire and their mechanical properties.
- explore the anatomical and functional development of the amnion, yolk sac, umbilical cord, and placenta.
We use an array of approaches including fixed and live imaging of embryos and tissue explants, omics on cell populations and micro-dissected structures.
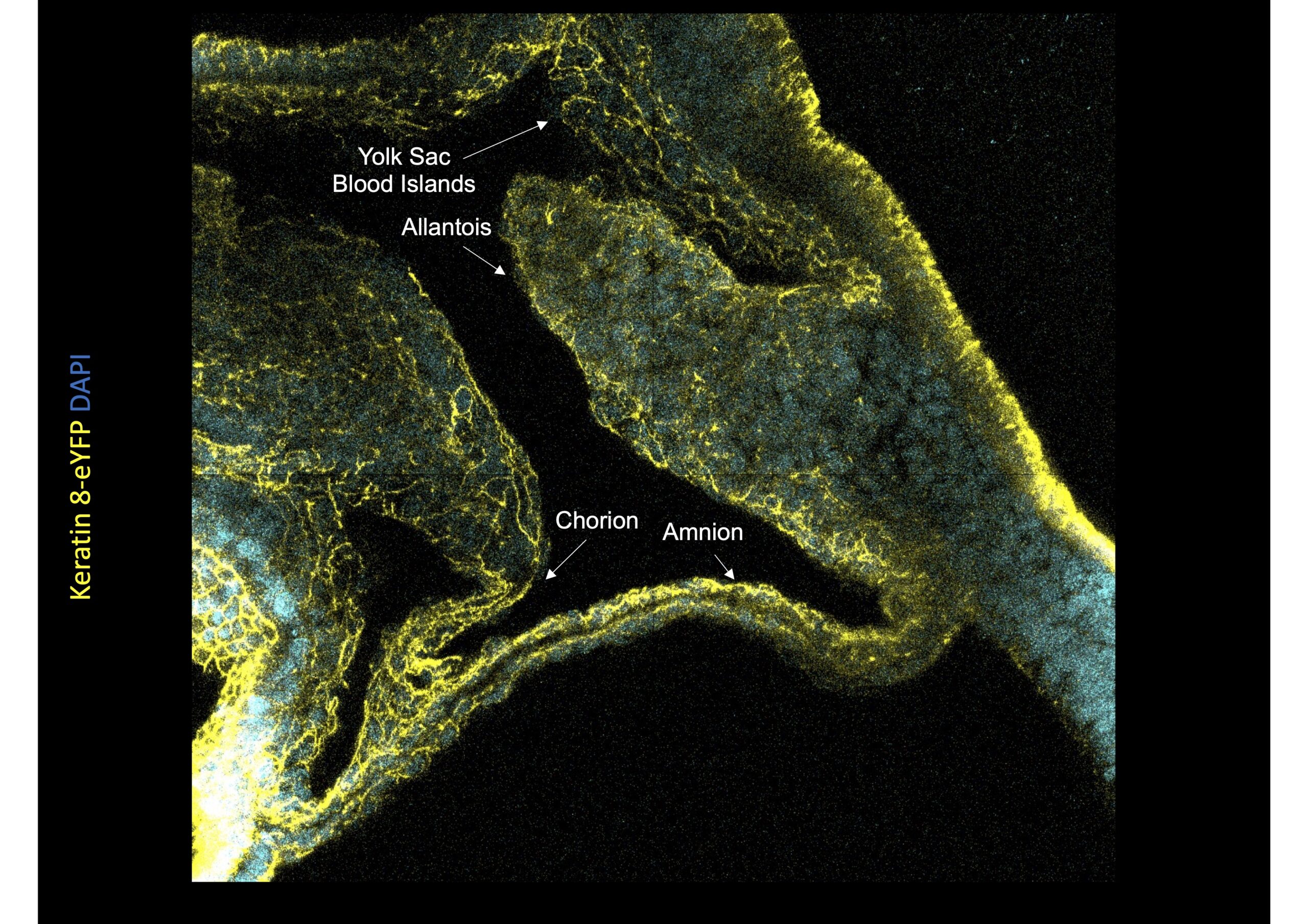
Nahaboo et al., EMBO 2022
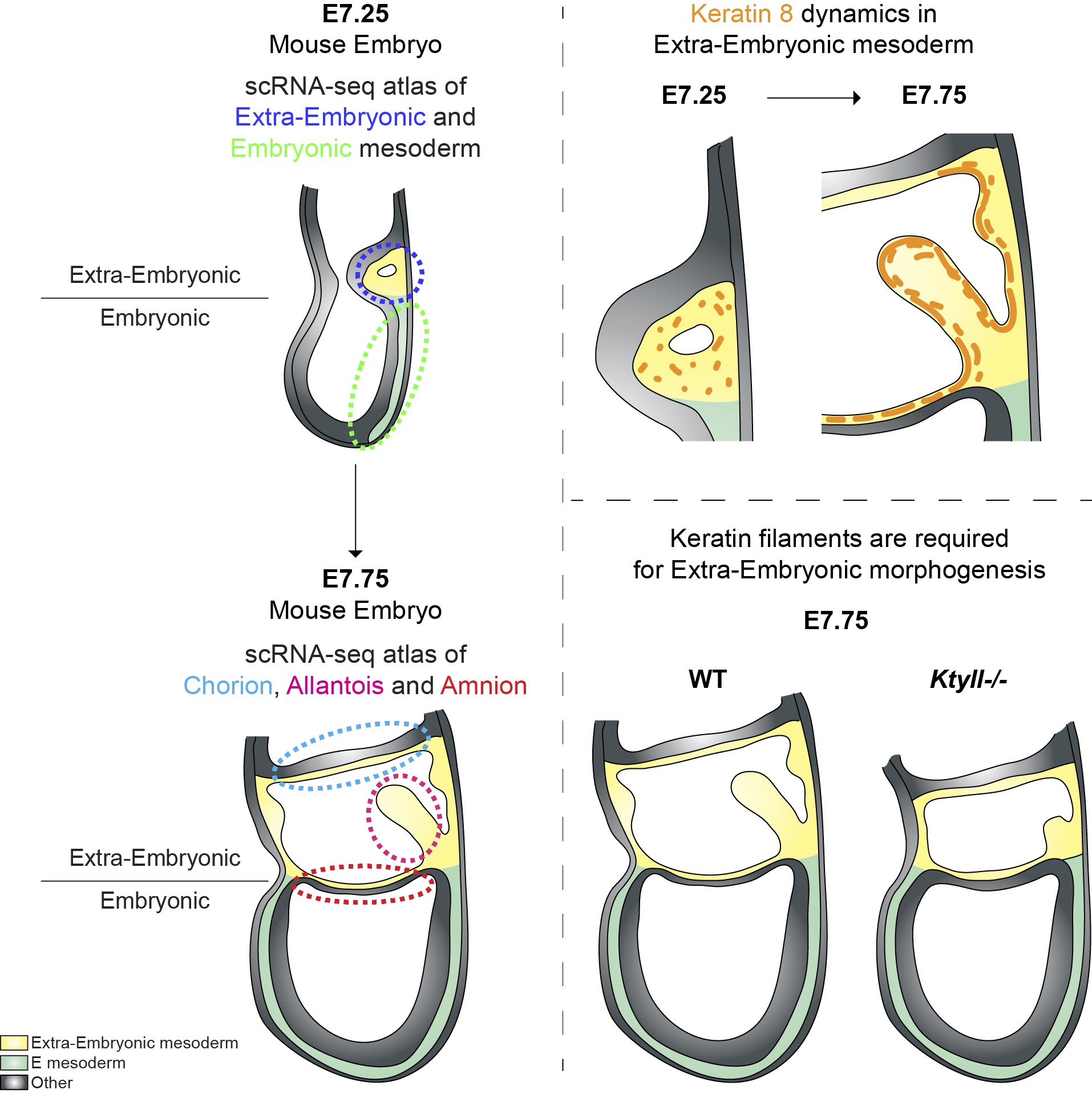
Nahaboo et al., EMBO 2022
Funding
FNRS/FRIA
Welbio
Fonds Erasme
Phoenix Erasmus
Fonds David et Alice van Buuren
Fondation Jaumotte-Demoulin
Group members
Isabelle Migeotte – Principal Investigator Research Associate F.N.R.S (isabelle.migeotte@ulb.be)
phone # +32 (0)2 555 4529

I received a MD PhD from Université Libre de Bruxelles, and certified in Internal Medicine and Medical Genetics. I trained in Developmental Biology with Kathryn Anderson in Sloan-Kettering Institute in NYC. I became a FNRS Research Associate and Principal Investigator at IRIBHM in 2012. My laboratory focuses on the cellular mechanisms regulating morphogenesis of the mouse embryo and its extra-embryonic membranes. In parallel, I teach genetics to bachelor students in medicine and biomedical sciences and am a consultant at the ULB Centre of Human Genetics, with clinical (medical genetics consults) and research (diagnostic techniques/ gene discovery in clinical cohorts) interests.
![]()
Evangéline Despin-Guitard, PhD Student

I received my master degree in Cell biology and Development from the Université Paul Sabatier (Toulouse, France) in 2018. I trained for my master thesis under the supervision of Eric Theveneau at the Center for Developmental Biology in Toulouse, were I studied epithelial to mesenchymal transition during neural crest delamination in the avian embryo. I joined the Migeotte Lab in January 2019 and obtained a FRIA/FNRS scholarship in October 2019. My work focuses on the relationship between cell cycle and epithelial to mesenchymal transition during gastrulation of the mouse embryo.
Eliana Nehme, PhD Student
 I graduated from Notre Dame University-Lebanon (NDU) with a Bachelor of Science degree majoring in Medical Laboratory Technology. Afterwards, I was granted a scholarship that enabled me to complete my master’s degree in Biomedical Sciences: Biochemistry and Molecular Genetics at the American University of Beirut-Lebanon (AUB). My work focused on the epigenetic suppression of a group of evolutionary conserved transcription factors, TBX2 subfamily (TBXs 2, 3, 4 and 5) in human non-small cell lung cancer (NSCLC). I occupied a position of medical laboratory training preceptor at NDU.
I graduated from Notre Dame University-Lebanon (NDU) with a Bachelor of Science degree majoring in Medical Laboratory Technology. Afterwards, I was granted a scholarship that enabled me to complete my master’s degree in Biomedical Sciences: Biochemistry and Molecular Genetics at the American University of Beirut-Lebanon (AUB). My work focused on the epigenetic suppression of a group of evolutionary conserved transcription factors, TBX2 subfamily (TBXs 2, 3, 4 and 5) in human non-small cell lung cancer (NSCLC). I occupied a position of medical laboratory training preceptor at NDU.
Passionate about research, I joined Isabelle Migeotte’s lab in November 2022. My work focuses on the molecular determinants for the mechanical properties of the mouse embryo extra-embryonic membranes.
![]()
Kristof Van Schoor, PhD Student
 I obtained a professional bachelor’s degree in medical laboratory technology from the University of Applied Sciences and Art in Ghent in 2018. For my bachelor’s thesis, I studied the effects of decalcification on immunohistochemical staining in bone marrow diagnostics. In 2022, I received a master’s degree in biomedical sciences from the KU Leuven. I wrote my master thesis on canonical BMP signalling in the development of the intestinal lymphatic system with Prof. An Zwijsen at the Center for Molecular and Vascular Biology (KU Leuven). In November 2022, I joined the IRIBHM to study the development of the extraembryonic vasculature and the attachment of the umbilical cord to the placenta
I obtained a professional bachelor’s degree in medical laboratory technology from the University of Applied Sciences and Art in Ghent in 2018. For my bachelor’s thesis, I studied the effects of decalcification on immunohistochemical staining in bone marrow diagnostics. In 2022, I received a master’s degree in biomedical sciences from the KU Leuven. I wrote my master thesis on canonical BMP signalling in the development of the intestinal lymphatic system with Prof. An Zwijsen at the Center for Molecular and Vascular Biology (KU Leuven). In November 2022, I joined the IRIBHM to study the development of the extraembryonic vasculature and the attachment of the umbilical cord to the placenta
![]()
Sorya Fagnoul, PhD Student
 After completing my medical training (ULB) in 2016, I specialized in nephrology in 2022. I joined the Migeotte Lab in October 2022 with a grant from the Fonds Erasme. My project will endeavour to improve diagnosis of kidney diseases of genetical eatiology. In addition to monogenic diseases, I will explore more complex modes of inheritance, such as oligenic diseases, thanks to a novel method created by our colleagues at IB² (Interuniversity Institute of Bioinformatics in Brussels). In kidney transplantation, I will evaluate the value of a genetic diagnosis to reduce renal risk for both donor and recipient, but also maximise their matching to avoid organ rejection. Finally, I aim to establish a platform of renal organoids to test the variants of interest we will identify.
After completing my medical training (ULB) in 2016, I specialized in nephrology in 2022. I joined the Migeotte Lab in October 2022 with a grant from the Fonds Erasme. My project will endeavour to improve diagnosis of kidney diseases of genetical eatiology. In addition to monogenic diseases, I will explore more complex modes of inheritance, such as oligenic diseases, thanks to a novel method created by our colleagues at IB² (Interuniversity Institute of Bioinformatics in Brussels). In kidney transplantation, I will evaluate the value of a genetic diagnosis to reduce renal risk for both donor and recipient, but also maximise their matching to avoid organ rejection. Finally, I aim to establish a platform of renal organoids to test the variants of interest we will identify.
![]()
Ayoub Radi, Lab technician
 I got my Diploma in Biotechnology from Institut Roger Lambion in June 2019. I started working as a lab technician at the IRIBHM in October 2019 in the group of Marc Parmentier until October 2022 where I joined Isabelle Migeotte’s lab. In parallel, I am also currently doing a Master in Industrial Engineering at the ISIPS.
I got my Diploma in Biotechnology from Institut Roger Lambion in June 2019. I started working as a lab technician at the IRIBHM in October 2019 in the group of Marc Parmentier until October 2022 where I joined Isabelle Migeotte’s lab. In parallel, I am also currently doing a Master in Industrial Engineering at the ISIPS.
![]()
Alumni:
Wallis Nahaboo, PhD
Latifa Hammou
Bechara Saykali, PhD
Marie-Lucie Racu, MD
Diana Suarez Boomgaard, PhD
Navrita Mathiah, PhD
Agathe Denys
Publications
Selected Publications
Van Schoor Kristof , Bruet Emmanuel , Jones Elizabeth Anne Vincent , Migeotte Isabelle. Origin and flow-mediated remodeling of the murine and human extraembryonic circulation systems, Frontiers in Physiology, Volume 15 – 2024 |https://doi.org/10.3389/fphys.2024.1395006
Evangéline Despin-Guitard, Ronan Quenec’Hdu, Wallis Nahaboo, Nicole Schwarz, Rudolf E.Leube, Claire Chazaud and Isabelle Migeotte. Frontiers in Cell and Developmental Biology (2022) “Regionally specific levels and patterns of keratin 8 expression in the mouse embryo visceral endoderm emerge upon anterior-posterior axis determination” Frontiers in Cell and Developmental Biology (2022) https://doi.org/10.3389/fcell.2022.1037041
Nahaboo W., Eski S.E., Vermeersch M., Saykali B., Monteyne D., Magin T., Schwarz N., Zwijsen A, Perez-Morga D., Singh S. P. Migeotte I. (2021) . “Keratin dynamics govern the establishment of the maternal-fetal interface”.
The EMBO Journal (2022). e108747.https://doi.org/10.15252/embj.2021108747
Nahaboo W, Saykali B, Mathiah N, Migeotte I. (2021) Visualizing Mouse Embryo Gastrulation Epithelial-Mesenchymal Transition Through Single Cell Labeling Followed by Ex Vivo Whole Embryo Live Imaging. Methods Mol Biol. 2021;2179:135-144. doi: 10.1007/978-1-0716-0779-4_12.PMID: 32939718
N. Mathiah, E. Despin-Guitard, M. Stower, W. Nahaboo, E. S. Eski, S. P. Singh, S. Srinivas, and I. Migeotte, “Asymmetry in the frequency and position of mitosis in the mouse embryo epiblast at gastrulation”, EMBO Rep, vol. 9, no. n/a, p. 317, Oct. 2020.
Q. Zhang et al. “Inborn errors of type I IFN immunity in patients with life-threatening COVID-19,” Science Sep. 2020, p. eabd4570. (Migeotte. I, IRIBHM)
Kyprianou, C., Christodoulou, N., Hamilton, R. S., Nahaboo, W., Suárez-Boomgaard, D., Amadei, G., Migeotte, I., & Zernicka-Goetz, M. Basement membrane remodelling regulates mouse embryogenesis. (2020) Nature (London), 582(7811), 253-258. doi:10.1038/s41586-020-2264-2
Delavallée, L., Mathiah, N., Cabon, L., Mazeraud, A., Brunelle-Navas, M.-N., Lerner,L. L., Tannoury, M., Prola, A., Moreno-Loshuertos, R., Baritaud, M., Vela, L., Garbin, K., Garnier, D., Lemaire, C., Langa-Vives,F., Cohen-Salmon, M., Fernández-Silva, P., Chretien, F., Migeotte, I., & Susin, S. A. Mitochondrial AIF loss causes metabolic reprogramming, caspase-independent cell death blockade, embryonic lethality, and perinatal hydrocephalus (2020). M. Molecular metabolism, 40, 101027. doi:10.1016/ j.molmet.2020.101027
Saykali, B., Nahaboo, W., Mathiah, N., Racu, M.-L., Defrance, M., & Migeotte, I. (2019). Distinct mesoderm migration phenotypes in extra-embryonic and embryonic regions of the early mouse embryo. eLife, 8, e42434. doi:10.7554/eLife.42434.001
Nahaboo, Wallis, and Migeotte, Isabelle (2018) Cleavage and Gastrulation in the Mouse Embryo. In: eLS. John Wiley & Sons Ltd, Chichester. http://www.els.net doi:10.1002/9780470015902.a0001068.pub3
Browet, A. A., De Vleeschouwer, C., Jacques, L. L., Mathiah, N., Saykali, B., & Migeotte, I. (2016). Cell segmentation with random ferns and graph-cuts. Proceedings – International Conference on Image Processing. 7533140, 4145-4149. doi:10.1109/ICIP.2016.7533140.
Mazari, E., Zhao, X., Migeotte, I., Collignon, J., Gosse, C., & Perea-Gomez, A. (2014). A microdevice to locally electroporate embryos with high efficiency and reduced cell damage. Development, , dev.106633. http://doi.org/10.1242/dev.106633
Zhao, X., Mazari, E., Suárez-Boomgaard, D. D., Migeotte, I., Perea-Gomez, A., & Gosse, C. (2014). Finite element model simulations to assist the design of microdevices dedicated to the localized electroporation of mouse embryos. E C S Transactions, 64(16), 7 14. doi:10.1149/06416.0007ecst
Simonis, N., Migeotte, I., Lambert, N., Perazzolo, C., de Silva, D. C., Dimitrov, B., Heinrichs, C., Janssens, S., Kerr, B., Mortier, G., Van Vliet, G., Lepage, P., Casimir, G., Abramowicz, M., Smits, G., & Vilain, C. (2013, June). FGFR1 mutations cause Hartsfield syndrome, the unique association of holoprosencephaly and ectrodactyly. Journal of medical genetics, 50(9), 585-592. doi:10.1136/jmedgenet-2013-101603
Bloomekatz, J., Grego-Bessa, J., Migeotte, I., & Anderson, K. V. (2012). Pten regulates collective cell migration during specification of the anterior-posterior axis of the mouse embryo. Developmental Biology, 364(2), 192–201. http://doi.org/10.1016/j.ydbio.2012.02.005
Migeotte, I., Grego-Bessa, J., & Anderson, K. V. (2011). Rac1 mediates morphogenetic responses to intercellular signals in the gastrulating mouse embryo. Development (Cambridge, England), 138(14), 3011–3020. http://doi.org/10.1242/dev.059766
Lee, J. D., Migeotte, I., & Anderson, K. V. (2010). Left-right patterning in the mouse requires Epb4.1l5-dependent morphogenesis of the node and midline. Developmental Biology, 346(2), 237–246. http://doi.org/10.1016/j.ydbio.2010.07.029
Migeotte, I., Omelchenko, T., Hall, A., & Anderson, K. V. (2010). Rac1-dependent collective cell migration is required for specification of the anterior-posterior body axis of the mouse. PLoS Biology, 8(8), 37–38. http://doi.org/10.1371/journal.pbio.1000442
Job opportunities