Our research
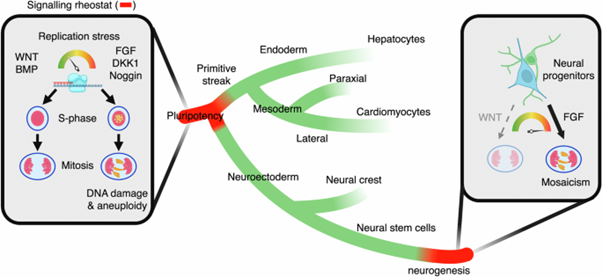
Embryonic development relies on genome and proteome stability in all species. Despite the robustness of the developmental programme, embryo fitness is continuously threatened by de novo acquisition of metabolic, proteostatic and genomic alterations in cells. While resilience to stress has been studied in several model organisms (including C. Elegans or D. Renio), it remains unexplored in mammals, and particularly in humans, in which spontaneous miscarriage is frequent during the first weeks of gestation4–6. While frequent gain or losses of chromosomes (aneuploidy) in some cells of the embryo are considered one of the known hallmarks of early pregnancy loss, we do not yet understand: i) what are the cellular causes of aneuploidy, and whether ii) microenvironmental stresses could lead to the high rates of embryo failure. Recently, we have identified that specific cellular signaling cascades triggers DNA replication stress in pluripotency, a process that fades down after specification of the three germ layers and re-emerges during neurogenesis (https://www.nature.com/articles/s41467-024-51821-9). This lineage-specific response to genotoxic stress makes us hypothesize whether a specific lineage response to diverse type of cellular stresses occurs during early development. (Figure 1A).
De Jaime-Soguero, et al. Nature Communications (2024)
Developmental resilience at the cellular level
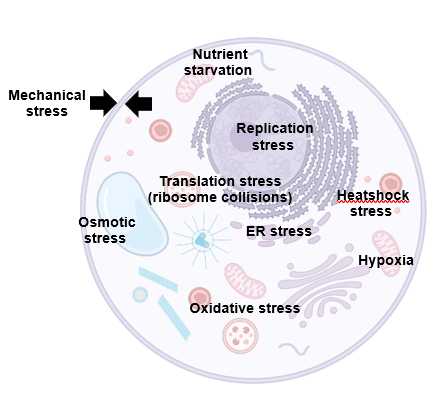 Among microenvironmental stresses, we are currently studying the impact of oxidative stress and protein folding stress in lineage-specific responses, as it has been previously associated with embryo failure through unknown molecular mechanisms we aim to decipher. Our lab aims to i) characterize the intrinsic physiological levels of diverse stressors (ie. ER stress, oxidative stress, heat-shock stress, hypoxia) across lineage specification and ii) study how extrinsic induction of the stressors shape cellular resilience and genome integrity through cell division in different mouse and human early lineages across space and time. Building on the laboratory experience using in vitro 2D and 3D human and mouse stem cell models (pluripotent stem cells lineage specification pipelines, gastruloids, EiTiX embryoids), the expertise in early mouse embryo development and our collaborators (Migeotte lab), and an array of classical and advanced molecular biology techniques (including single-cell sequencing), our lab investigates i) how mammalian lineages mitigate/respond/adapt to specific stress cues, and ii) the transcriptional and metabolic response, fate and chromosome stability in the context of cellular stress, providing ultimately a proof of concept of the developmental bottlenecks at risk in early human development and shedding light on the causes of spontaneous miscarriage.
Among microenvironmental stresses, we are currently studying the impact of oxidative stress and protein folding stress in lineage-specific responses, as it has been previously associated with embryo failure through unknown molecular mechanisms we aim to decipher. Our lab aims to i) characterize the intrinsic physiological levels of diverse stressors (ie. ER stress, oxidative stress, heat-shock stress, hypoxia) across lineage specification and ii) study how extrinsic induction of the stressors shape cellular resilience and genome integrity through cell division in different mouse and human early lineages across space and time. Building on the laboratory experience using in vitro 2D and 3D human and mouse stem cell models (pluripotent stem cells lineage specification pipelines, gastruloids, EiTiX embryoids), the expertise in early mouse embryo development and our collaborators (Migeotte lab), and an array of classical and advanced molecular biology techniques (including single-cell sequencing), our lab investigates i) how mammalian lineages mitigate/respond/adapt to specific stress cues, and ii) the transcriptional and metabolic response, fate and chromosome stability in the context of cellular stress, providing ultimately a proof of concept of the developmental bottlenecks at risk in early human development and shedding light on the causes of spontaneous miscarriage.
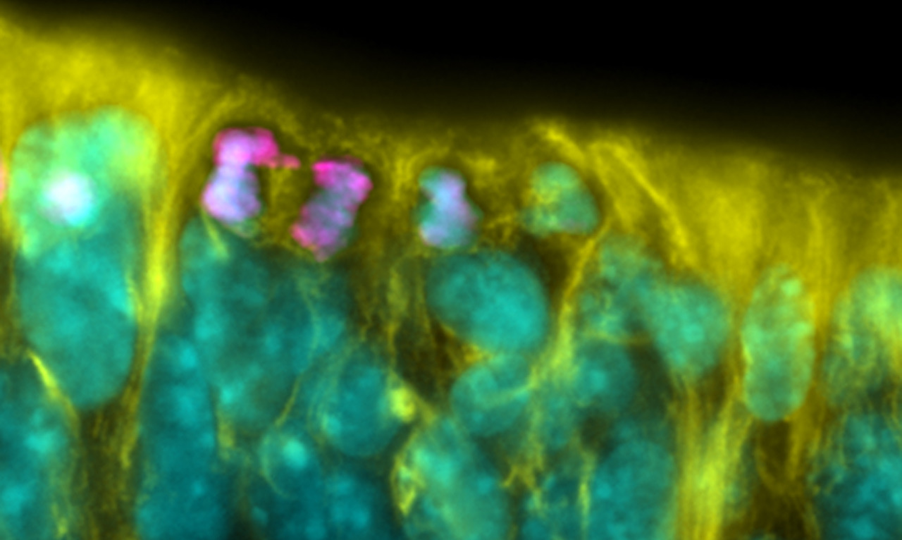
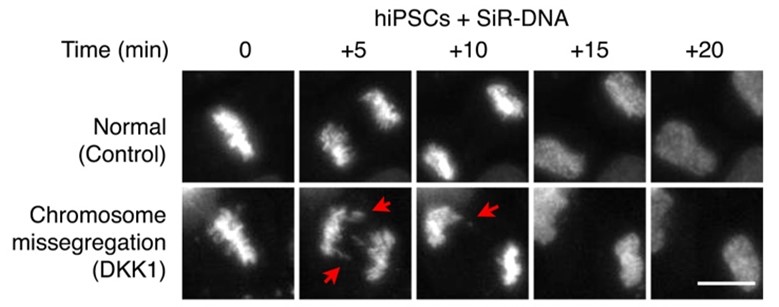
Chromosome segregation fidelity in human pluripotency
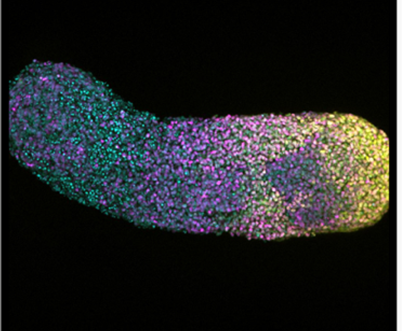
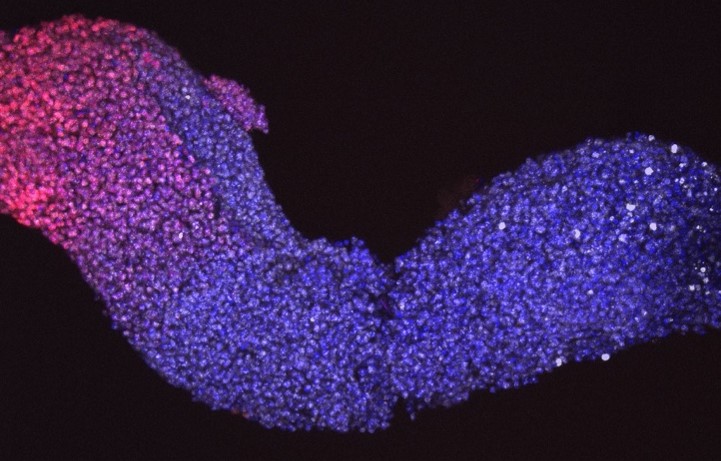
Human gastruloid derived from hiPSCs (120h after aggregation)
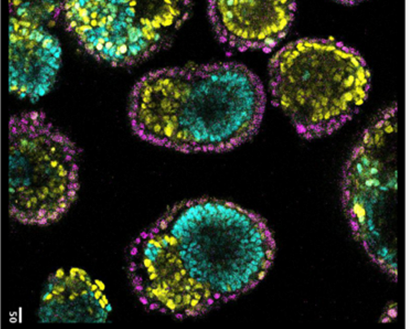
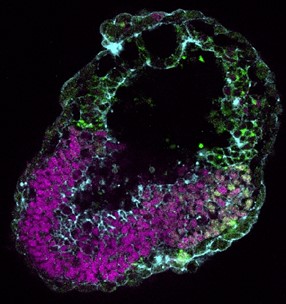
Mouse EiTiX embryoid generated from mouse embryonic stem cells (Day 5 after aggregation)
Group members

Anchel de Jaime Soguero – Junior group leader (anchel.de.jaime.soguero@ulb.be)
I received my PhD from KU Leuven (Belgium), where I studied the transcriptional regulation of Wnt signaling. In 2019, I moved to the University of Heidelberg under the guidance of Prof. Sergio Acebron where I unveiled how morphogenetic signals regulate genome integrity and chromosome stability. Since October 2024, I lead my group at IRIBHM dissecting cellular stress responses across lineage states and mammalian embryo development.
Ana-Irina Ivanova – MSc student
“I completed my Bachelor’s and Master’s degrees in Biomedical Sciences at the Universite Libre de Bruxelles (ULB). Since October 2024, I have been conducting my Master’s thesis research in the lab, focusing on the impact of oxidative stress in mouse pluripotency and lineage specification.”
Publications
Selected publications
De Jaime-Soguero*, J. Hattemer*, A. Bufe, A. Haas, J. van den Berg, V. van Batenburg, B. Das, B. di Marco, S. Androulaki, N. Bohly, J.J.M. Landry, B. Schoell, V.S. Rosa, L. Villacorta, Y. Baskan, M. Trapp, V. Benes, A. Chabes, M. Shahbazi, A. Jauch, U. Engel, A. Patrizi, R. Sotillo, A. van Oudenaarden, J. Bageritz, J. Alfonso, H. Bastians, and S.P. Acebron. Developmental signals control chromosome segregation fidelity during pluripotency and neurogenesis by modulating replicative stress. Nature Communications (2024). 15, 7404 (2024). https://doi.org/10.1038/s41467-024-51821-9
Van den Berg, V. van Batenburg, C. Geisenberger, R. Tjeerdsma, A. de Jaime-Soguero, S.P. Acebron, M.A.T.M. Van Gut, A. van Oudenaarden. Quantifying DNA replication speeds in single cells by scEdU-seq. Nature Methods (2024) 21, 1175–1184. https://doi.org/10.1038/s41592-024-02308-4
Athanasouli*, M. Balli*, A.de Jaime-Soguero#, A. Boel, S. Papanikolaou, B.K. van der Veer, A. Janiszewski, T. Vanhessche, A. Francis, Y.E. Laithy, A.L. Nigro, F. Aulicino, K.P. Koh, V. Pasque, M. Pia Cosma, C. Verfaillie, A. Zwijsen, B. Heindryckx, C. Nikolaou & F. Lluis#. The Wnt/TCF7L1 transcriptional repressor axis drives primitive endoderm formation by antagonizing naive and formative pluripotency. Nature Communications (2023) 14, 1210. https://doi.org/10.1038/s41467-023-36914-1 (#co-corresponding author)
Giebel*, A. de JaimeSoguero*, A. García-del Arco, J.J.M. Landry, M. Tietje, L. Villacorta, V. Benes, V. Fernandez-Saiz, S.P. Acebron. USP42 protects ZNRF3/RNF43 from R-spondin-dependent clearance and inhibits Wnt signalling. EMBO Reports (2021) 22 (5), e51415. https://doi.org/10.15252/embr.202051415 (*co-first author)
Bufe, A. García-del Arco, M. Hennecke, A. de Jaime-Soguero, M. Ostermaier, Y.C. Lin, A. Ciprianidis, J. Hattemer, U. Engel, P. Beli, H. Bastians, S.P. Acebron. Wnt signaling recruits KIF2A to the spindle to ensure chromosome alignment during mitosis. PNAS (2021) 118 (34), e2108145118. https://doi.org/10.1073/pnas.2108145118
Job offers


Doctoral Fellow – Cellular and developmental biology
Genome and proteome integrity are crucial for proper embryo development, but how both are monitored across lineage specification remains an open question. We have recently identified that morphogenetic and patterning signals regulate DNA replication dynamics and chromosome segregation fidelity in a lineage-specific manner (De Jaime-Soguero et al. Nature Communications, 2024; PMID 39191776). These findings suggest that specific lineages respond and mitigate differently replicative stress. We seek a motivated doctoral researcher to investigate developmental resilience during early embryogenesis and lineage specification towards other stress sources. Our recently formed group (De Jaime-Soguero lab, IRIBHM-ULB) has expertise in cellular signaling and quantification of genome stability, pluripotency and early lineage specification in 2D, novel 3D stem cell-based embryo models and ex vivo mouse embryo experimentation. We collaborate with Isabelle Migeotte lab at IRIBHM, which has long-standing expertise in gastrulation and whole-embryo live imaging, and with bioinformaticians at the home institute and at EMBL Gene Core (Heidelberg).
Qualification profile
You hold a MsC in Life Sciences or Bioinformatics and have a strong interest in molecular, cell or developmental biology. Experience with cloning and/or R coding for bioinformatic analysis is a plus. You have strong analytical and communication skills, are enthusiastic and critical thinker. Fluency in English is required, as well as to be able to work well in an international environment. Grades during the MsC will be taken in account during the selection.
We offer
A 1-year funded PhD contract with the possibility of extension. During this time, the candidate is expected to apply for PhD fellowship programs under the support of IRIBHM. Our laboratory offers: i) a healthy lab culture and lively international environment, with English as working language, ii) access to advanced training in scientific and professional skills, iii) supervision and support tailored to your career perspectives, iv) access to state-of-the-art core facilities at ULB (microscopy, flow cytometry, sequencing) and v) interdisciplinary collaborations involved.
Why to join IRIBHM and come to live in Brussels?
IRIBHM-J.E. Dumont is a multidisciplinary research institute at the Université Libre de Bruxelles (ULB, Belgium) with a focus in diverse topics such as signal transduction, development, neuroscience, cancer and computational biology (https://iribhm.org ; our group page is under construction). At the heart of Europe, Brussels is the largest student city in Belgium welcoming 160 different nationalities, a vibrant, tolerant and international capital.
How to apply:
Send your application to Anchel de Jaime-Soguero (dejaimesoguerolab@gmail.com), with the subject lane: De Jaime-Soguero lab PhD application. Attach: (1) Your BsC and MsC transcripts, (2) a detailed CV, (3) the contact information of two previous mentors/supervisors, and (4) a cover letter stating your interest in our lab and project. Applications will be evaluated on a rolling basis, until the position is filled. Applications sent after 31th of January 2025 will not be considered. The starting date expected is March 2025 onwards (to be discussed).