Our research
The computational biology group at IRIBHM focuses on the transcriptomes of, mostly but not exclusively, breast and thyroid cancers. Two major topics are currently investigated.
A-to-I editing by the enzyme ADAR substitutes inosines for adenosines at specific positions in double stranded RNAs (dsRNA) and can very substantially alter a cell’s transcriptome and function. While investigation of RNA editing in cancer is still in its infancy, recent studies point to an important role in cancer. We have shown that, overall, A-to-I editing is a pervasive, yet reproducible, source of variation that is globally controlled by amplification of chromosome and inflammation, both of which are highly prevalent among human cancers. Importantly, ADAR expression also modulates the proliferation and apoptosis of cancer cells. We currently investigate RNA editing at the level of noncoding RNA, its intratumoral heterogeneity and its relation with inflammation in cancer.
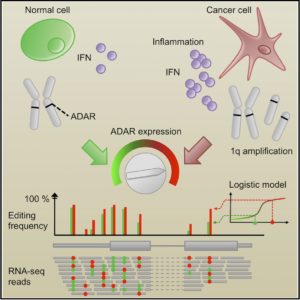
Principles governing RNA editing in cancer.
Textbooks suggest that cancers are compact balls with an inner core and an invasive front in contact with non-cancerous cells, and that tumor expansion is driven by tumor cells proliferation subsequently to oncogenic mutations in their genomes. We recently reconstructed at histological scale the 3D volume occupied by tumor cells in a BRAFV600E-mutated thyroid cancer with low tumor purity, as determined by sequencing, but initially considered high purity during pathology review.
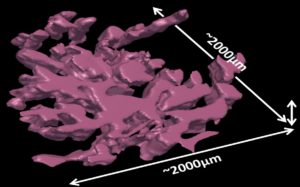
3D reconstruction of a thyroid tumor at histological scale.
In contrast with a compact ball, tumor cells form a sparse mesh deeply embedded within the stroma. The concepts of inner core and invasive front brakes down in this morphology: all tumor cells were within short distance from the stroma. The fibrous stroma was highly cellular and proliferative. This case is not unique: 3.5% of thyroid cancers have purities <25%. Moreover, a substantial fraction of BRAFV600E tumors are associated with extensive fibrosis, high stromal activation, and dedifferentiation.
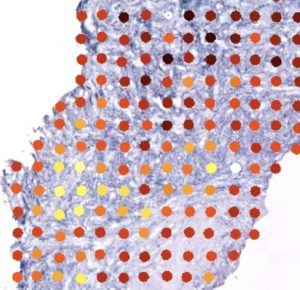
Spatially-resolved transcriptome of a thyroid tumor overlaid onto the corresponding histological image.
We are currently in the process of developing this line of research about intra tumoral heterogeneity with single cell RNA-seq and by implementing at IRIBHM a spatial transcriptomics, which makes it possible to resolve the transcriptome at thousands of locations along the three spatial dimensions of tumor blocks.
Group members
Vincent Detours (head) (Vincent.Detours@ulb.ac.be)
phone # +32(0)2 555 4220
Helen Libion (PhD student)
Joël Rodrigues-Vitoria (Phd Student)
Ruben Lattuca (Phd Student)
Adrien Tourneur (PhD student)
Publications
Peer-reviewed publications (* equal contributors; # corresponding author)
Kyrilli A., Gacquer D., Detours V., Lefort A., Libert F., Twyffels L., Van den Eeckhaute L., Strickaert A., Maenhaut C., De Deken X., Dumont J.E., Miot F., Corvilain B., (2019), Dissecting the role of thyrotropin in the DNA damage response in human thyrocytes after 131I, γ radiation and H2O2, J. Clin. Endocrinol. Metab., 10.1210/clinem/dgz185.
Solinas C., Marcoux D., Garaud S., Vitória J.R., Van den Eynden G., de Wind A., De Silva P., Boisson A., Craciun L., Larsimont D., Piccart-Gebhart M., Detours V., t’Kint de Roodenbeke D., Willard-Gallo K. (2019), BRCA gene mutations do not shape the extent and organization of tumor infiltrating lymphocytes in triple negative breast cancer. Cancer Lett. 450:88-97, doi: 10.1016/j.canlet.2019.02.027
Ghaddhab C, Kyrilli A, Driessens N, Van Den Eeckhaute E, Hancisse O, De Deken X, Dumont JE, Detours V, Miot F, Corvilain B. (2019), Factors contributing to the resistance of the thyrocyte to hydrogen peroxide, Mol Cell Endocrinol., 481:62-70, doi: 10.1016/j.mce.2018.11.010
Suzuki I.K., Gacquer D., Van Heurck R., Kumar D., Wojno M., Bilheu A., Herpoel A., Lambert N., Chéron J., Polleux F., Detours V., Vanderhaeghen P., (2018), Human-specific NOTCH2 paralogs expand human cortical neurogenesis through regulation of Delta/Notch interactions. Cell 31;173(6):1370-1384, doi: 10.1016/j.cell.2018.03.067. [Comments on this paper by E. Pennisi Science (2018) doi: 10.1126/science.360.6392.951; Nature May 31st 2018; A. Onela, The Scientist May 31st 2018; etc. News media coverage: New York Times, The Economist, Spiegle, Le Soir, Het Last Neuws, VRT, etc.]
Fimereli D., Fumagalli D., Brown D., Gacquer D., Rothé F., Salgado R., Larsimont D., Sotiriou C., Detours V., (2018), Genomic hotspots but few recurrent fusion genes in breast cancer. Genes Chromosomes Cancer. doi: 10.1002/gcc.22533.
Tarabichi M., Antoniou A., Le Pennec S., Gacquer D., de Saint Aubain N., Craciun L., Cielen T., Laios I., Larsimont D., Andry G., Dumont J.E., Maenhaut C., Detours V., (2018), Distinctive desmoplastic 3D morphology associated with BRAFV600E in papillary thyroid cancers, J Clin Endocrinol Metab. doi:10.1210/jc.2017-02279.
Solinas C., Garaud S., De Silva P., Boisson A., Van den Eynden G., de Wind A., Risso P., Rodrigues Vitória J., Richard F., Migliori E., Noël G., Duvillier H., Craciun L., Veys I., Awada A., Detours V. Larsimont D., Piccart-Gebhart M., Willard-Gallo K., (2017), Immune Checkpoint Molecules on Tumor-Infiltrating Lymphocytes and Their Association with Tertiary Lymphoid Structures in Human Breast Cancer. Front Immunol., doi:10.3389/fimmu.2017.01412.
Tarabichi M. & Detours V. (2016), A research note regarding “Variation in cancer risk among tissues can be explained by the number of stem cell divisions”, F1000 Research, doi:10.12688/f1000research.9448.1
Saiselet M., Pita J.M., Augenlicht A., Dom G., Tarabichi M., Fimereli D., Dumont JE., Detours V., Maenhaut C. (2016), miRNA expression and function in thyroid carcinomas: a comparative and critical analysis and a model for other cancers, Oncotarget, doi: 10.18632/oncotarget.9655.
Handkiewicz-Junak D., Swierniak M., Rusinek D., Oczko-Wojciechowska M., Dom G., Maenhaut C., Unger K., Detours V., Bogdanova T., Thomas G., Likhtarov I., Jaksik R., Kowalska M., Chmielik E., Jarzab M., Swierniak A. & Jarzab B., (2016), Gene signature of the post-Chernobyl papillary thyroid cancer, Eur J Nucl Med Mol Imaging, 2016 Jan 26.
Saiselet M., Gacquer D., Spinette A., Craciun L., Decaussin-Petrucci M., Andry G., Detours# V. & Maenhaut# C., (2015), New global analysis of the microRNA transcriptome of primary tumors and lymph node metastases of papillary thyroid cancer, BMC Genomics,16:828. doi: 10.1186/s12864-015-2082-3
Fumagalli F., Gacquer D. Rothé F, Lefort A., Libert F., Brown D., Kheddoumi N., Shlien A., Konopka, T, Salgado R., Larsimont D., Polyak K., Willard-Gallo K., Desmedt C., Piccart M., Abramowicz M., Campbell P.J., Sotiriou S. & Detours V., (2015), Principles Governing A-to-I RNA Editing in the Breast Cancer Transcriptome, Cell Reports, doi:10.1016/j.celrep.2015.09.032. [Comments on this paper by L. Shipman, Nat. Rev. Cancer (2015), doi:10.1038/nrc4044 ; Rayon-Estrada et al., Trends in Cancer (2015), doi:10.1016/j.trecan.2015.10.008]
Tarabichi M., Saiselet M., Trésallet C., Hoang C., Larsimont D., Andry G., Maenhaut C. & Detours V., Revisiting the transcriptional analysis of primary tumours and associated nodal metastases with enhanced biological and statistical controls: application to thyroid cancer, (2015), Br J Cancer. 112(10):1665-1674.
Fimereli D., Gacquer D., Fumagalli D., Salgado R., Rothé F., Larsimont D., Sotiriou C. & Detours V., (2015), No significant viral transcription detected in whole breast cancer transcriptomes, BMC Cancer 15(1):1176.
Le Pennec S., Konopka T., Gacquer D., Fimereli D., Tarabichi M., Tomás G., Savagner F., Decaussin-Petrucci M., Trésallet C., Andry G., Larsimont D., Detours# V. & Maenhaut# C., (2015), Intratumor heterogeneity and clonal evolution in an aggressive papillary thyroid cancer and matched metastases, Endocr. Relat. Cancer 22(2):205-16.
Fumagalli D., Blanchet-Cohen A., Brown D., Desmedt C., Gacquer D., Michiels S., Rothé F., Majjaj S., Salgado R., Larsimont D., Ignatiadis M., Maetens M., Piccart M. & Detours V., Sotiriou C., Haibe-Kains B., (2014), Transfer of clinically relevant gene expression signatures in breast cancer: from Affymetrix microarray to Illumina RNA-Sequencing technology, BMC Genomics 21;15(1):1008
Versteyhe S., Driessens N., Ghaddhab C., Tarabichi M., Hoste C., Dumont J.E., Miot F., Corvilain B. & Detours V., (2013), Comparative analysis of the thyrocytes and T-cells responses to H2O2 and radiation reveals an H2O2-induced antioxidant transcriptional program in thyrocytes, J. Clin. Endocrin. Metab., doi:10.1210/jc.2013-1266.
Tarabichi M., Antoniou A., Saiselet M., Pita J.M., Andry G., Dumont J.E., Detours V. & Maenhaut C., (2013), Systems biology of cancer: entropy, disorder, and selection-driven evolution to independence, invasion and “swarm intelligence”. Cancer Metastasis Rev. 32(3-4):403-21. doi: 10.1007/s10555-013-9431-y
Fimereli D., Detours V.# & Konopka T., (2013), TriageTools: tools for partitioning and prioritizing analysis of high-throughput sequencing data. Nucleic Acids Res., 41(7):e86. doi: 10.1093/nar/gkt094.
Tarabichi M., Detours V.# & Konopka T., (2012), Piecewise polynomial representations of genomic tracks. PLoS One, 7(11):e48941.
Coletta A., Molter C., Duque R., Steenhoff D., Taminau J., de Schaetzen V., Meganck S., Lazar C., Venet D., Detours V., Nowe A., Bersini H. & Weiss Solis D.Y., (2012), InSilico DB genomic datasets hub: an efficient starting point for analyzing genome-wide studies in GenePattern, Integrative Genomics Viewer, and R/Bioconductor, Genome Biol. 13(11) R104.
Venet D., Detours V. & Bersini H., (2012). A Measure of the Signal-to-Noise Ratio of Microarray, Samples and Studies Using Gene Correlations. PLoS One 7(12): e51013.
Dom G., Tarabichi M., Unger K., Thomas G., Oczko-Wojciechowska M., Bogdanova T., Jarzab B., Dumont J.E., Detours V. & Maenhaut C., (2012), A gene expression signature distinguishes normal tissues of sporadic and radiation-induced papillary thyroid carcinomas. Br. J. Cancer, doi:10.1038/bjc.2012.302.
Tomás G., Tarabichi M., Gacquer D., Hébrant A., Dom G., Dumont J.E., Keutgen X., Fahey T.J. 3rd, Maenhaut C. & Detours V., (2012), A general method to derive robust organ-specific gene expression-based differentiation indices: application to thyroid cancer diagnostic, Oncogene, Jan 23. doi:10.1038/onc.2011.626.
Detours V., (2011), Confounded Cancer Markers, The Scientist, December 7th 2011.
Venet D., Dumont J.E. & Detours V., (2011), Most Random Gene Expression Signatures are Significantly Associated with Breast Cancer Outcome, PLoS Computational Biology 7(10), e1002240. [Comments on this paper by Ng et al. (2012), Breast Cancer Res. 14, p313-5 and Jordan B. (2012) Bioassay 34(9), p730-3].
Lambert N., Lambot M.A., Bilheu A., Albert V., Englert Y., Libert F., Noel J.C., Sotiriou C., Holloway A.K., Pollard K.S., Detours V. & Vanderhaeghen P. (2011), Genes Expressed in Specific Areas of the Human Fetal Cerebral Cortex Display Distinct Patterns of evolution, PLoS One 6(3), e17753.
Maenhaut C., Detours V., Dom G., Handkiewicz-Junak D., Oczko-Wojciechowska M. & Jarzab B. (2011), Gene Expression Profiles of Radiation-induced Thyroid Cancers, Clin. Oncol. 23(4), p282-8.
van Staveren*, W.C.G., Weiss Solís*, D.Y., Hébrant, A., Detours, V., Dumont, J.E. & Maenhaut, C., (2009), Human cancer cell lines: Experimental models for cancer cells in situ? For cancer stem cells? BBA Cancer 1795(2), 92-103.
Detours, V., Versteyhe, S., Dumont J. E. & Maenhaut C., (2008), Gene Expression Profiles of Post-Chernobyl Thyroid Cancers, Curr. Op. in Endocrinol., Metabolism and Diabetes 15, p440-5.
28. Burniat, A., Jin, L., Detours, V., Driessens, N., Goffard, J.C., Santoro, M., Rothstein, J., Dumont, J.E., Miot, F. & Corvilain B., (2008), Gene Expression in RET/PTC3 and E7 Transgenic Mouse Thyroids: RET/PTC3 but not E7 Tumors are Partial and Transient Models of Human Papillary Thyroid Cancers. Endocrinology 149, p5107-17.
Detours, V., Delys, L., Libert, F., Weiss Solís, D., Bogdanova, T., Dumont, J. E., Franc, B., Thomas, G. & Maenhaut, C., (2007), Genome-wide Gene Expression Profiling Suggests Distinct Radiation Susceptibilities in Sporadic and Post-Chernobyl Papillary Thyroid Cancers, British J. Cancer 97, 818-25.
van Staveren* W.C., Weiss Solís* D.Y., Delys L., Duprez L., Andry G., Franc B., Thomas G., Libert F., Dumont J.E., Detours V., Maenhaut C., (2007), Human thyroid tumor cell lines derived from different tumor types present a common dedifferentiated phenotype, Cancer Res. 67(17):8113-20.
Delys*, L., Detours*, V., Franc, B., Thomas, G., Bogdanova, Tronko, M., T., Libert, F., Dumont, J. E. & Maenhaut, C., (2007), Gene expression and the biological phenotype of papillary thyroid carcinomas, Oncogene 26(57), 7894-903.
Song Y, Driessens N, Costa M, De Deken X, Detours V, Corvilain B, Maenhaut C, Miot F, Van Sande J, Many MC, Dumont JE, (2007), Roles of hydrogen peroxide in thyroid physiology and disease, J Clin Endocrinol Metab. 92(10), 3764-73.
van Staveren, C. G., Detours, V., Dumont, J. E., & Maenhaut, C., (2006), Negative feedbacks in normal cell growth and their suppression in tumorigenesis. Cell Cycle 5, 571-572.
van Staveren, C. G., Weiss, D., Delys, L., Venet, D., Cappello, M., Andry, G., Dumont, J. E., Libert, F., Detours, V., & Maenhaut, C., (2006), Gene Expression in TSH-treated Human Thyrocytes and Autonomous Adenomas: Suppression of Negative Feedbacks in Tumorigenesis, Proc. Nat. Acad. Sci. USA 103, 413–418
Detours, V., Dumont, J. E., & Maenhaut, C., (2006), Systems Biology, Cell Specificity and Physiology, in Theories in Medicine, Paton R. & McNamara L. Eds. Elsevier.
Wattel, S., Mircescu, H., Venet, D., Burniat, A., Franc, B., Frank, S., Andry, G., Van Sande, J., Rocmans, P., Dumont, J. E., Detours, V. & Maenhaut, C., (2005), Gene Expression in Thyroid Autonomous Adenomas Provides Insight on their Physiopathology, Oncogene 24, 6902-16.
Detours, V., Wattel, S., Venet, D., Hutsebaut, N., Bogdanova, T., Tronko, M. D., Dumont, J. E., Franc, B., Thomas, G. & Maenhaut, C., (2005), Absence of a Specific Radiation Signature in Post-Chernobyl Thyroid Cancers, British J. Cancer 92, 1545-1552.
Detours, V., Dumont, J. E., Bersini, H., & Maenhaut, C., (2003). Integration and Cross-Validation of High-Throughput Gene Expression Data: Comparing Heterogenous Data Sets, FEBS Lett. 546, 98-102.
Yusim, K., Kesmir, C., Addo, M. M., Altfeld, M., Gaschen, B., Chigaev, A., Detours, V. & Korber, B. T., (2002), Clustering Patterns of CTL Epitopes in HIV-1 Proteins Reveal Imprints of Immune Evasion on HIV-1 Global Variation, J. Virology, 76(17), 8757-8768.40. Kesmir, C. Nussbaum, N. K., Shild, H., Detours, V., Brunak, S., (2002), Prediction of Proteasome Clivage Motifs by Neural Networks, Protein Engeneering, 15(4), 287-296.
Korber, B. T., Gaschen, B., Yusim, K., Thakallapally, R., Kesmir, C. & Detours, V., (2001), Evolutionary and Immunological Implications of Contemporary HIV-1 Variation, Brit. Med. Bull., 58, 19-42.
Detours, V. & Perelson, A. S., (2000), The Paradox of Alloreactivity and Self MHC Restriction: Quantitative Analysis and Statistics, Proc. Nat. Acad. Sci. USA, 97, 8479-8483.
Detours, V., Mehr, R., & Perelson, A. S., (2000), Deriving Quantitative Constraints Under Which T Cell Selection Operates from Data on the Mature T Cell Repertoire, J. Immunol. 164, 121-128.
Detours, V., Mehr, R., & Perelson, A. S., (1999) A Quantitative Theory of Affinity-Driven T Cell Repertoire Selection, J. theor. Biol. 200, 389-403.
Detours, V., & Perelson, A. S., (1999), Explaining High Alloreactivity as a Quantitative Consequence of Affinity-Driven T Cell Selection, Proc. Nat. Acad. Sci. USA 96, 5153-5158.
Detours, V., Sulzer, B., & Perelson A. S., (1996), Size and Connectivity of the Idiotypic Network are Independent of the Discreteness of the Affinity Distribution, J. theor. Biol. 183, 408-416.
Bersini, H., & Detours, V. #, (1994), Asynchrony Induces Stability in Cellular Automata Based Models, in Proceedings of the Fourth Conference on Artificial Life, Brooks, R. A., Maes, P., Eds. MIT Press.
Detours,V. #, Bersini, H., Stewart, J., Varela, F. J., (1994), Development of Idiotypic Network in Shape Space, J. theor. Biol. 170, 401-414.